
September 22, 2025
Searching for the active compound in a haystack: Fabian Liessmann has successfully defended his doctoral thesis

In Jonathan Safran Foer’s book “Extremely Loud and Incredibly Close”, nine-year-old Oskar Schell embarks on a journey through New York. Oskar is in search of the right lock for a small key he found in his late father’s possession. However, his quest is about more than solving a mystery; it becomes a journey of self-discovery, insight and understanding. Fabian Liessmann embarked on a similar journey as part of his doctoral studies, which he has now successfully completed. However, instead of searching for a metal lock, his goal was to discover new active compounds as drug candidates to combat diseases. Using modern computer programs and artificial intelligence, Fabian Liessmann searched through billions of possible substances.
Where the search for new drugs to combat diseases begins
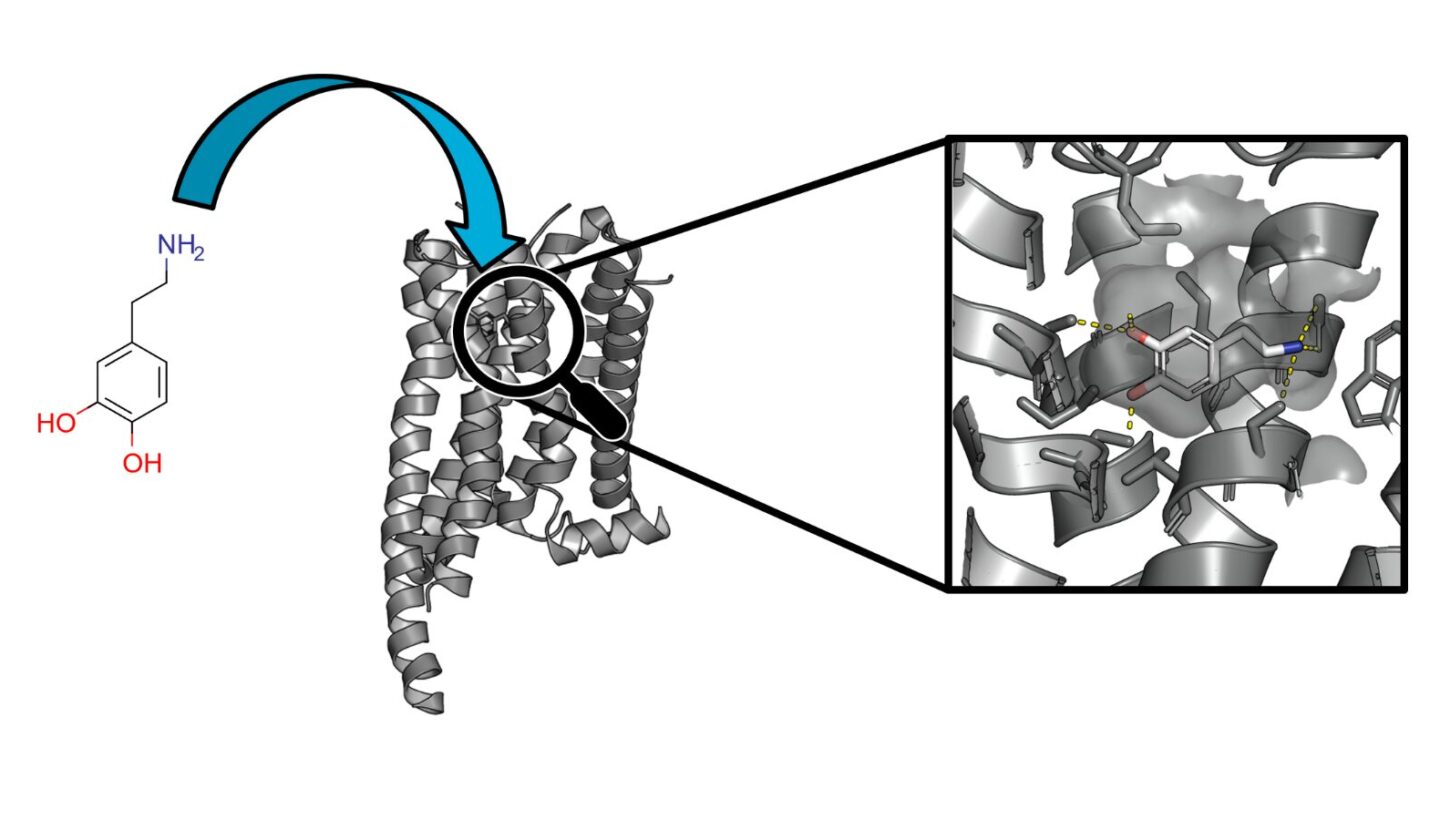
In this case, it takes us to a tiny building block of the human body: G protein-coupled receptors (GPCRs). These proteins act as switches in our cell membranes. They control numerous biological processes by receiving signals from the environment and transmitting them to the interior of the cell. Therefore, GPCRs are of great medical importance. Research has shown that up to one-third of all approved drugs act via these ‘switches’ by specifically activating, blocking or regulating their activity. However, we are far from understanding all of these receptors in the human body.
Small molecules can function as drugs against diseases and medical conditions such as obesity, Alzheimer’s disease and cancer. Scientists expect, that there are around 1060 of potential drug-like small molecules. Traditional methods require a lot of time and money to test even a few thousand of these molecules to identify novel drug candidates.

Artificial intelligence and algorithms for accelerating drug development
This is where Fabian Liessmann’s research comes in. In his doctoral thesis “Application and Optimization of Ultra-Large Library Screening for the Identification of Novel Small Molecule Ligands for G Protein-Coupled Receptors”, supervised by Prof. Jens Meiler, he investigated how researchers can use computers to search specifically for new active compounds that bind well to such G protein-coupled receptors. Rather than testing millions of substances in the laboratory, ultra-large library screening uses various algorithms, artificial intelligence and 3D models of receptors to test billions of virtual molecules on a computer. Fabian Liessmann has developed methods and workflows that predict whether a substance will fit well into the binding pocket of a G protein-coupled receptor, much like a key fitting into a lock. This enables new drugs to be developed more quickly and cheaply. And in a more targeted manner, even for receptors that have hardly been researched to date.
“I found the idea particularly exciting that somewhere in the vast, largely unexplored chemical space, there are active compounds against diseases that we don’t even know about yet. The possibility of using modern algorithms and artificial intelligence to analyze billions of such molecules on a computer instead of laboriously testing them in the laboratory immediately fascinated me.”
Fabian Liessmann
Fabian Liessmann began his doctorate under Prof. Jens Meiler at the Institute for Drug Discovery at Leipzig University and ScaDS.AI Dresden/Leipzig. Before, he studied pharmacy at Leipzig University. Alongside research stays at Vanderbilt University in the US, he co-founded AI-Driven Therapeutics in October 2024 with other researchers from his group. The company accelerates protein design using computer-aided AI methods. He successfully defended his dissertation on June 27th, 2025.






