June 6, 2025
New AI Method Improves Transcript Detection in Microscopy Images

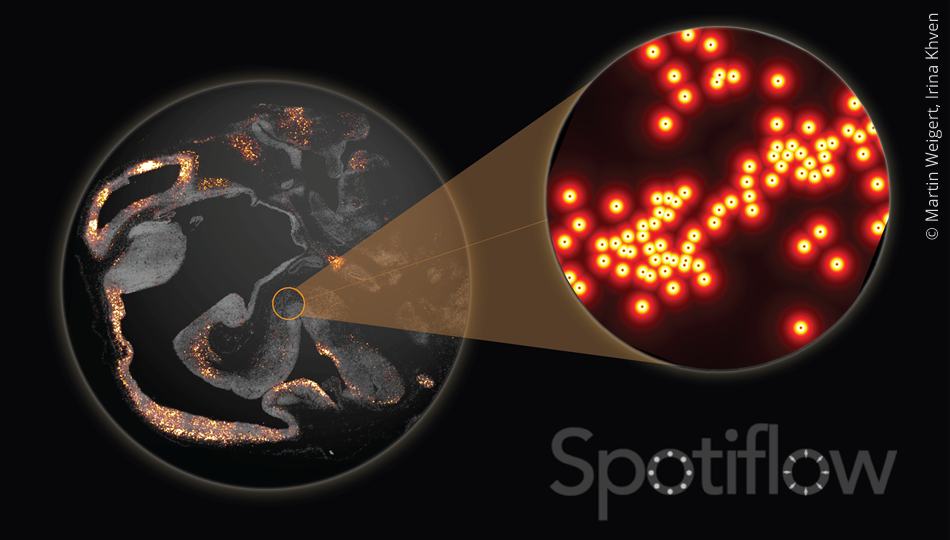
Understanding gene activity within tissues requires analyzing 2D and 3D microscopy images to detect millions of transcript molecules appearing as tiny spots. This presents major challenges due to massive data scale and often challenging imaging conditions with current methods struggling with the accuracy and efficiency needed for reliable biological insights.
Researchers at TU Dresden / ScaDS.AI Dresden/Leipzig and École Polytechnique Fédérale de Lausanne (EPFL) have now developed Spotiflow, an AI method that improves large-scale transcript localization by combining deep learning with a novel geometric detection approach: Spotiflow formulates localization as a stereographic flow regression task, a geometric embedding technique that maps transcript locations into a specialized coordinate system, enabling more precise localization than traditional methods. Published in Nature Methods, Spotiflow demonstrates significant performance improvements over existing methods while being faster, and more memory-efficient. The AI system works across both 2D and 3D imaging conditions without requiring dataset-specific parameter adjustments, streamlining large-scale transcript analysis for high-throughput biological studies.
“This project is a great example of collaborative work between computer science and life sciences,” said Prof. Martin Weigert, senior author and professor at TU Dresden and ScaDS.AI Dresden/Leipzig. Working with the group of Prof. Gioele La Manno (EPFL), they combined computational expertise with biological understanding to address transcript detection as a major bottleneck in spatial transcriptomics pipelines. By bringing AI into the equation, first author Albert Dominguez Mantes developed a robust and scalable method that works without complex parameter tuning to make advanced transcript detection accessible to researchers across different areas.
Spotiflow is available as open-source Python package and Napari plugin.






