Computational B-cell epitope prediction using artificial intelligence methods
Title: Computational B-cell epitope prediction using artificial intelligence methods
Project duration: 01/23 – 12/24
Research Area: Life Science and Medicine
We are developing machine learning methods to predict B cell epitopes taking the conformational change of viral glycoproteins into account. AxIEM (Accelerated class I fusion protein Epitope Mapping) is a method to identify possible B-cell epitope regions on viral glycoproteins (especially class I fusion glycoproteins) which are recognized by the humoral immune response, especially, by antibodies. Determining additional epitope regions on the glycoproteins is necessary to inform rational vaccine design.
The methods that we are developing requires two different conformations of the glycoprotein. Unfortunately, there are only few determined structures. We will overcome by leveraging AlphaFold2-Multimer, which is a deep learning protein structure prediction tool- to extend the benchmark set. Together with an experimental scientist, we aim to test our predicted epitopes in the laboratory.
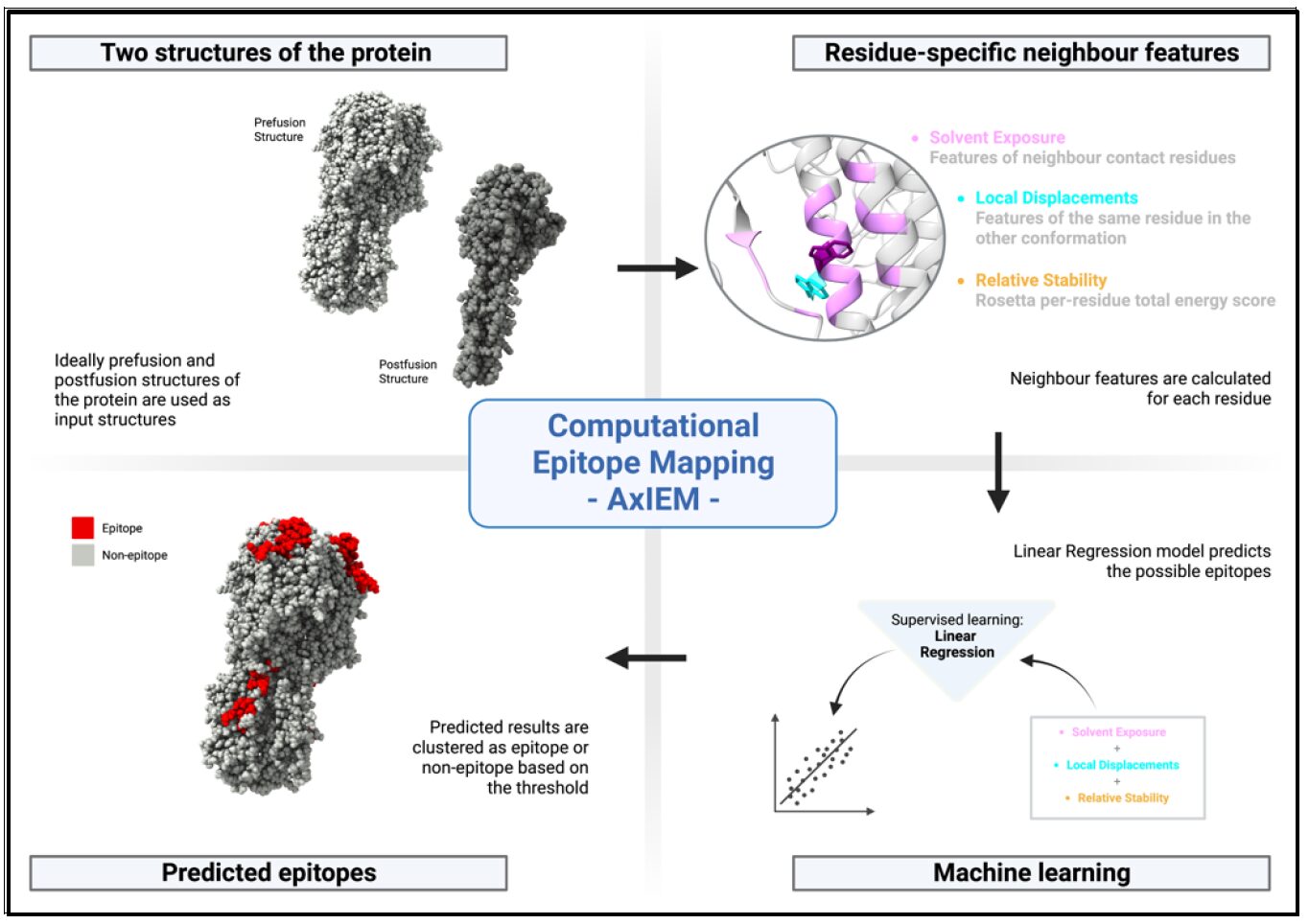
Aims
- optimize the AxIEM method for general use in epitope prediction of class I fusion
- proteins
- extend the current benchmark set of the AxIEM to predict B-cell epitopes for all class
I fusion proteins from AlphaFold predicted structures - test AxIEM in a real-world setting using the MARV glycoprotein as test case
- develop a new AI-driven protocol for conformational states prediction
Problems
Developing new antibodies and vaccines by only experimental methods is not time efficient and still very expensive. We hypothesize that computational methods can help to develop rational vaccines in short time and increase the efficiency of the experiments.
Practical Example
We predicted possible epitope regions for Marburg virus and based on the current result we
are optimizing and atomizing the protocol.
Technology
We use AlphaFold2-multimer which is the deep learning method to predict the structure of the protein. To predict B-cell epitopes of class I fusion glycoproteins, we optimize the AxIEM method which is also machine learning-based method.
Outlook
The project will support to develop rational vaccine with machine learning-based methods.
Team
Lead
- Jun.-Prof. Dr. Clara T. Schoeder
Team Members
- Sevilay Gülesen
Partners
- Prof. Dr. Jens Meiler
- Felipe Engelberger
- Johanna Möller



