July 2, 2024
Mutation Explorer – A 3D Tool for Protein Mutation Visualization and Analysis

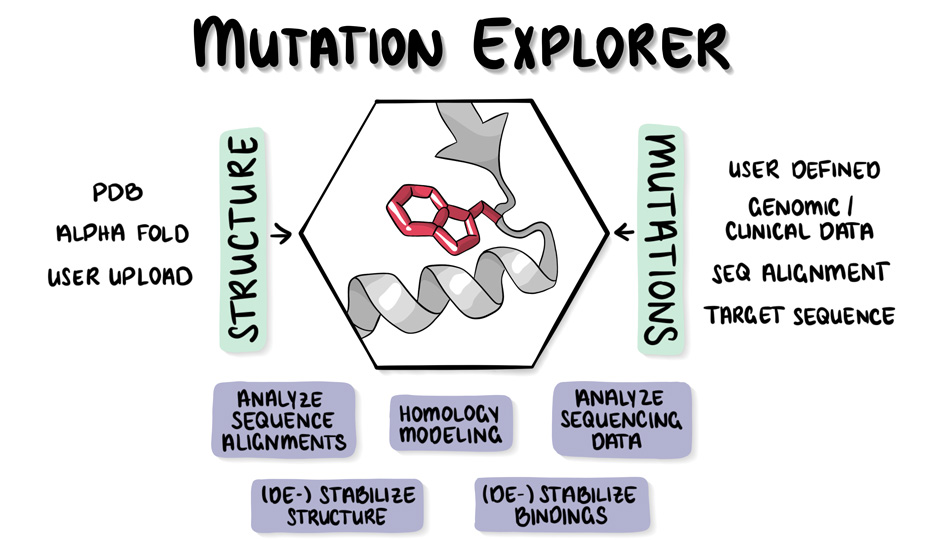
It all boils down to answering one question: How does a mutation in a protein contribute to an observed or desired phenotype? A better understanding of these processes will help to address questions about drug resistance in bacteria, genetic diseases in humans, and the development of cancer cells. Accessible and user-friendly applications for predicting, exploring, and visualizing protein complexes are extremely useful for studying mutations and their effects on these complexes. Current sequencing methods are very successful in identifying and revealing the risks of phenotype effects. However, they often struggle to explain the underlying molecular mechanisms. This is where 3D-Visualization tools, such as the new Mutation Explorer, co-developed by ScaDS.AI Dresden/Leipzig, can help.
AI-Driven Web Server for Protein Mutation and Stability Analysis
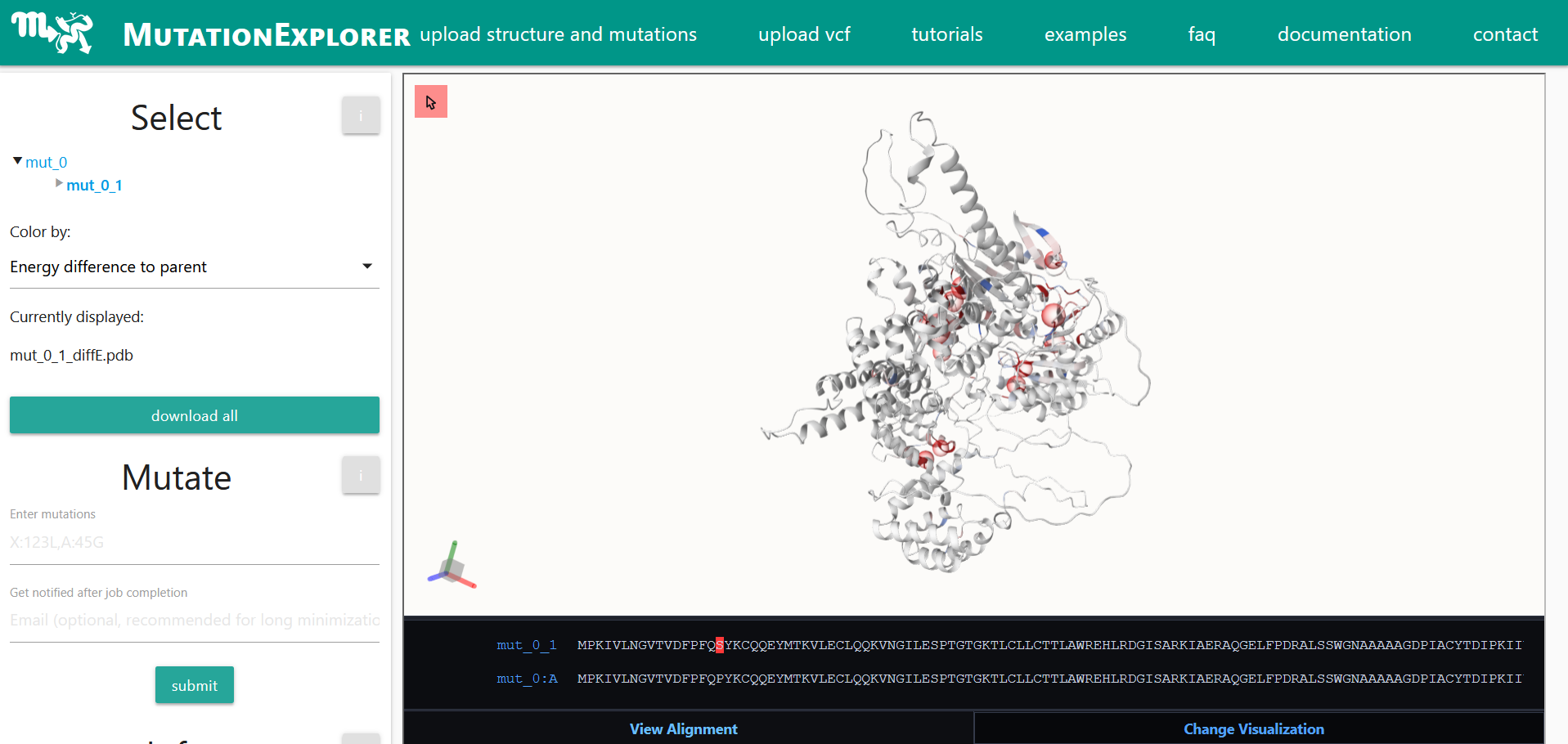
Mutation Explorer is a web server that allows users to mutate protein models from the Protein Data Base (PDB) via web interface. The effects of these mutations can then be analyzed and studied using several integrated tools. For example, protein structures can be observed through multiple rounds of mutations. Human SNPs (single nucleotide polymorphisms) can also be uploaded to visualize the destabilizing effects of mutations on molecular composition. Through a variety of interactive visualization options, users can quickly and intuitively understand the energetic changes that occur with mutations. These changes then affect the stability of the entire protein. Specifically, MutationExplorer uses an AI model that predicts the extent to which a cancer-related mutation, for example, can change the stability of the protein. This assists the user in their selection and analysis process.
Overall, the web server aims to provide an intuitive and deep understanding of disease-relevant genetic mutations. In addition to AI, the server also incorporates statistical and physics-based methods to further improve the simulation. For high-quality and accurate results, the simulation places special emphasis on energetic minimization in the protein structure. The smaller, the more accurate. This energetic stability of a mutated protein is calculated by the structure prediction program Rosetta, pioneered by Prof. Jens Meiler (Institute for Drug Design).
Collaborative Effort in Protein Structure Research
Mutation Explorer is part of a broader collaboration between Prof. Peter Hildebrand (Institute of Medical Physics and Biophysics) and Dr. Daniel Wiegreffe (part of Prof. Gerik Scheuermann’s group, Institute of Computer Science). Their collaboration promotes structure-based research in biochemistry and biophysics by developing and providing user-friendly and interactive online visualization tools ([1],[2]).
For the development of MutationExplorer, three teams from ScaDS.AI Dresden/Leipzig collaborated internationally. This collaboration included Leipzig University in Germany, the University of Copenhagen in Denmark, Vanderbilt University in Nashville, USA, and the University Hospital of Bielefeld University in Germany. Funding support is provided by the Federal Ministry of Education and Research of Germany and the Sächsische Staatsministerium für Wissenschaft, Kultur und Tourismus, under project identification number ScaDS.AI Dresden/Leipzig.






