Computational peptide design to target therapy-resistant tumors using physics- and machine learning-based methods
Title: Computational peptide design to target therapy-resistant tumors using physics- and machine learning-based methods
Project duration: 10.2023 – 06.2025
Research Area: Life Science and Medicine
Chronic lymphocytic leukemia (CLL) is one of the most common adult cancers of the body’s white blood cells, called lymphocytes, and is characterized by the accumulation of cells that are functionally incompetent to fight infection. One of the most commonly used treatments for CLL is a combination of chemotherapy and an anti-CD20 monoclonal antibody, such as rituximab. [2] The efficacy of rituximab in cancer treatment is based in part on its ability to induce complement-dependent cytotoxicity (CDC) to destroy target cells.
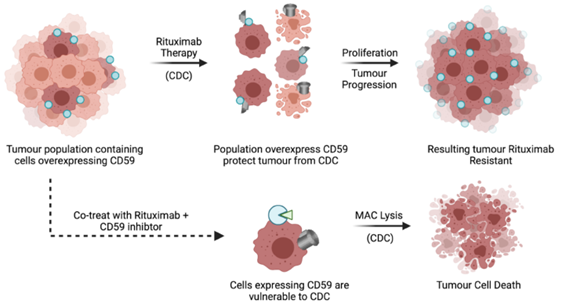
Human CD59 (hCD59) plays a critical role as a complement regulatory protein by limiting the assembly of the pore-forming membrane attack complex, thereby preventing activation of CDC. Lymphoma cells that overexpress CD59 directly contribute to increased resistance to rituximab-induced CDC therapy. [3] The development of bispecific CD20-CD59 antibodies or co-treatment with a CD59 inhibitor shows improved efficacy in complement-dependent cytotoxicity (see also Figure 1). [4] Enhancing mAb therapies such as rituximab with novel adjuvants, e.g. peptide inhibitors, to increase lytic efficiency may improve clinical outcomes and patient survival in CLL.
Aims
- Rational design of CD59 peptide binders to inhibit the interactions with the pore-forming membrane attack complex and, therefore, inducing cell death
- Developing a general pipeline for creating and designing de-novo cyclic peptide binders for arbitrary targets
- Utilization and extension of state-of-the-art physics-based and machine learning-based frameworks for the generation of cyclic peptides
- Integration of AI and biophysical design strategies
- Extending the method of cyclic peptide design to viral glycoproteins such as the Marburg virus
Problems
Developing new therapeutic peptides using only a wet chemistry laboratory is expensive, labor-intensive, and requires deep expertise. In contrast, computational drug design allows new potential drug candidates to be generated in silico using well-established design methods and frameworks. A prominent recent example of this opportunity is the design and evaluation of a new therapeutic compound targeting the spike protein for the treatment of COVID-19. [5] For this project, we want to design cyclic peptide inhibitors because they have better biological activity than their linear counterparts and are resistant to hydrolysis. However, machine learning methods for cyclic peptides are still underrepresented.
Practical Example
We are currently testing an extension of the two deep learning methods Alphafold and RFdiffusion. RFdiffusion, a generative network that generates protein backbones from random noise, was modified to generate cyclic backbones. Sequences were generated using the deep learning method ProteinMPNN, and an in-silico experiment is currently underway to benchmark the combination of these methods on known cyclic peptide backbones from the Protein Data Bank (PDB). After in-silico validation with docking studies, the method will be extended to CD59.
Technology
We employ machine learning-based tools for protein structure prediction and de-novo generation, including AlphaFold2 and RFdiffusion. In computational protein design, we use deep learning techniques like ProteinMPNN and physics-based methods like Rosetta.
Publications
- [1] Couves, E. (2022). “Disentangling the structural and cellular driving forces of CD59 inhibition of Membrane Attack Complex pore formation.” [Doctoral dissertation, Imperial College London].
- [2] Brown, Jennifer R et al. “The Role of Rituximab in Chronic Lymphocytic Leukemia Treatment and the Potential Utility of Biosimilars.” The oncologist vol. 23,3 (2018): 288-296. doi:10.1634/theoncologist.2017-0150
- [3] Hu, Weiguo et al. “Human CD59 inhibitor sensitizes rituximab-resistant lymphoma cells to complement-mediated cytolysis.” Cancer research vol. 71,6 (2011): 2298-307. doi:10.1158/0008-5472.CAN-10-3016
- [4] Stadlbauer, K. et al. “Bispecific mAb2 Antibodies Targeting CD59 Enhance the Complement- Dependent Cytotoxicity Mediated by Rituximab”. Int. J. Mol. Sci. 2022, 23, 5208. https://doi.org/10.3390/ijms23095208
- [5] Sarmadi, Soroush et al. “In Silico Design and Evaluation of a Novel Therapeutic Agent Against the Spike Protein as a Novel Treatment Strategy for COVID-19 Treatment.” Recent patents on biotechnology vol. 18,2 (2024): 162-176. doi:10.2174/1872208317666230523105759
Team
Lead
- Prof. Dr. Jens Meiler (Leipzig University)
Team Members
- Max Beining (Leipzig University)
Partners
- Jun.-Prof. Dr. Christina Lamers (Leipzig University)
- Jun.-Prof. Dr. Clara T. Schoeder (Leipzig University)




