Mapping Single-Cell Data between Species by Combined Deep Learning and Explainable AI
Title: Mapping Single-Cell Data between Species by Combined Deep Learning and Explainable AI
Project duration: 01.01.2022 – 30.06.2025
Research Area: Life Science and Medicine, Medical AI
In the project “Mapping single-cell data between species by combined deep learning and explainable AI”, we develop a principled approach to compare the cell-type- and disease-state specific molecular response between human disease conditions and corresponding animal models at the single-cell RNA sequencing (scRNAseq) transcriptome level. We apply our approach in two contexts: First, we map molecular states of whole blood samples between human COVID-19 patients and two hamster species developing moderate (Mesocricetus auratus) or severe disease course (Phodopus roborovskii) following SARS-CoV-2 infection. Second, we focus on side-effects following immunomodulatory therapies. We identify corresponding states of immune-related toxicities between cynomolgus monkeys (Macaca fascicularis) and humans in peripheral blood mononuclear cell subpopulations (PBMCs).
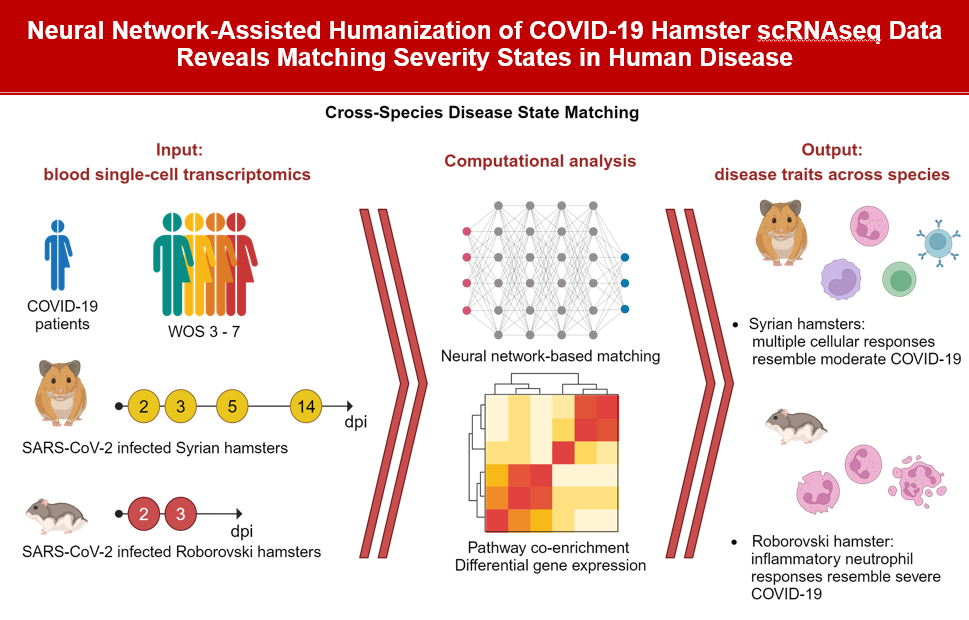
Aims
- transcriptome-based quantification of disease state similarities across species
- integrating deep learning with interpretable approaches
- establishing a respective robust, general methodologic framework
- improving the transferability of pre-clinical animal models towards human disease conditions
Problems
- technological and biological batch effects to be considered
- sparse, noisy, and high-dimensional data
- challenging biological interpretation
Technology
- single-cell RNA-sequencing analysis
- deep learning, variational autoencoder
- empirical Bayes moderation
Outlook
- Extension to other diseases and conditions for preclinical research and drug development
- Method developments to improve explainability of findings
Publications
- preprint: DOI: 10.1101/2024.01.11.574849v1
Team
Lead
- Markus Scholz (IMISE)
- Geraldine Nouailles (Charité)
- Kristin Reiche (Fraunhofer IZI),
Team Members
- Holger Kirsten (IMISE)
- Martin Witzenrath (Charité)
- Vincent David Friedrich
- Peter Pennitz (Charité)
Partners


