Computational protein design for novel immunotherapeutic drugs using artificial intelligence
Title: Computational protein design for novel immunotherapeutic drugs using artificial intelligence
Project duration: 10.2023 – 12.2024
Research Area: Life Science and Medicine
We are using and developing machine learning approaches for the rational design of proteins, yielding proteins of increased stability, with novel functionalities or altered binding properties to therapeutic targets. The computationally predicted constructs are then evaluated in the laboratory to translate the theoretical predictions into real-world data.
We are developing vaccine candidates for emerging viral infections and pandemic threats, protein therapeutics for cancer immunotherapy, and peptide and small molecule binders to modulate therapeutic protein targets.
The combination of rapidly evolving machine learning solutions for the prediction of protein structure and protein design together with our biomolecular methods for the evaluation of designed compounds allows us to develop novel drugs rationally and swiftly for pressing therapeutic needs.
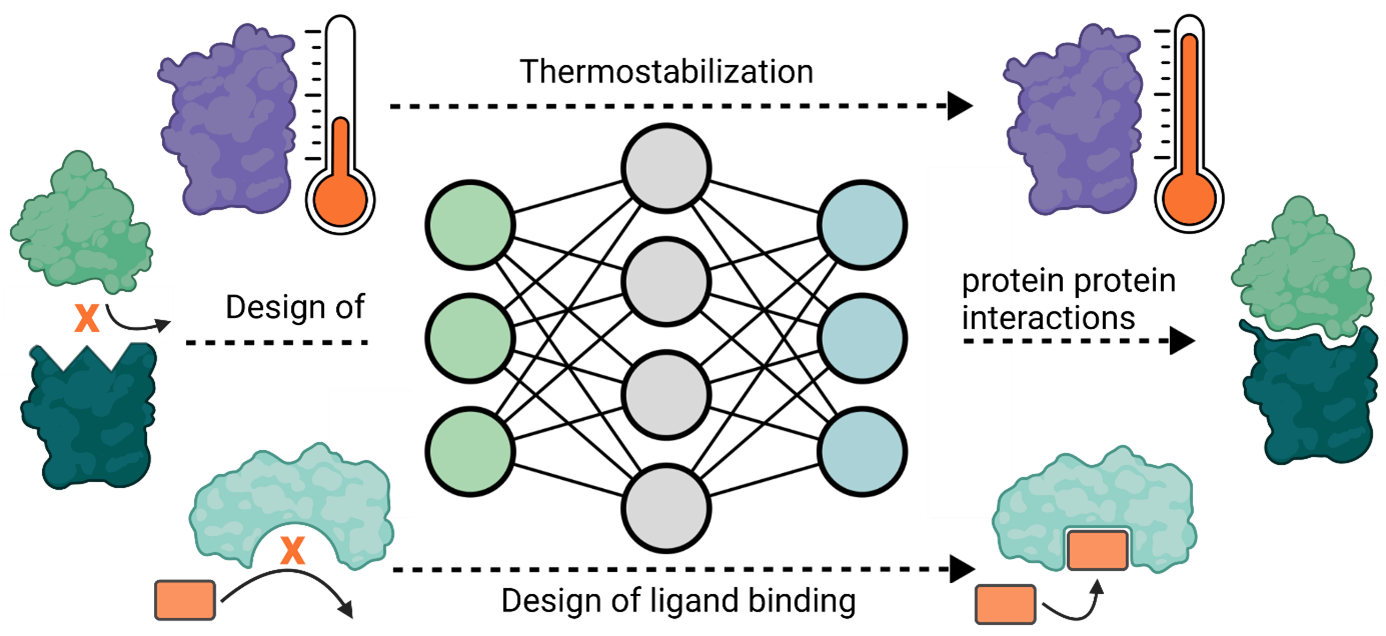
Aims
- Rational design of vaccine candidates for emerging viral infections to increase pandemic preparedness
- Development of stabilized single-chain T cell receptors as building blocks in anti-tumor and anti-viral therapies
- New methods for AI-driven protein design
- Integration of AI and biophysical design strategies
Problems
The development of new immunotherapeutic drugs is expensive, labor-intensive and requires expert knowledge. We hypothesize that AI-based protein design strategies will be able to counteract this and help to generate protein drugs more efficiently.
Practical Example
Currently, we are testing an AI-method for the design of hyperthermostable proteins, which will be useful for biotechnological applications. Also, we are generating interfaces for AI methods in classic protein design software frameworks.
These technologies are actively tested in our laboratories in the Institute for Drug Discovery.
Technology
We use machine learning-based protein structure prediction tools like AlphaFold2 and RFdiffusion. Computational protein design is carried out with deep learning-based methods for sequence design, like ProteinMPNN, but also Rosetta, leveraging its physics- and statistics-based energy function for the evaluation of designed structures.
Outlook
Our projects will be the foundation for AI-designed new biotechnological drugs.
Team
Lead
- Jun.-Prof. Clara T. Schoeder (Leipzig University)
Team Members
- Johanna Möller (Leipzig University)
- Moritz Ertelt (Leipzig University)



